Escherichia coli (E. coli) is a pathogenic bacteria which continues to be a predominant cause of infection in most countries worldwide with implications such as severe diarrhea, and urinary tract infection and even life-threatening neonatal meningitis. Antibiotic resistance of such bacteria is growing and therefore many conventional methods of managing the infections are becoming less effective, necessitating the development of novel means of combating infections.
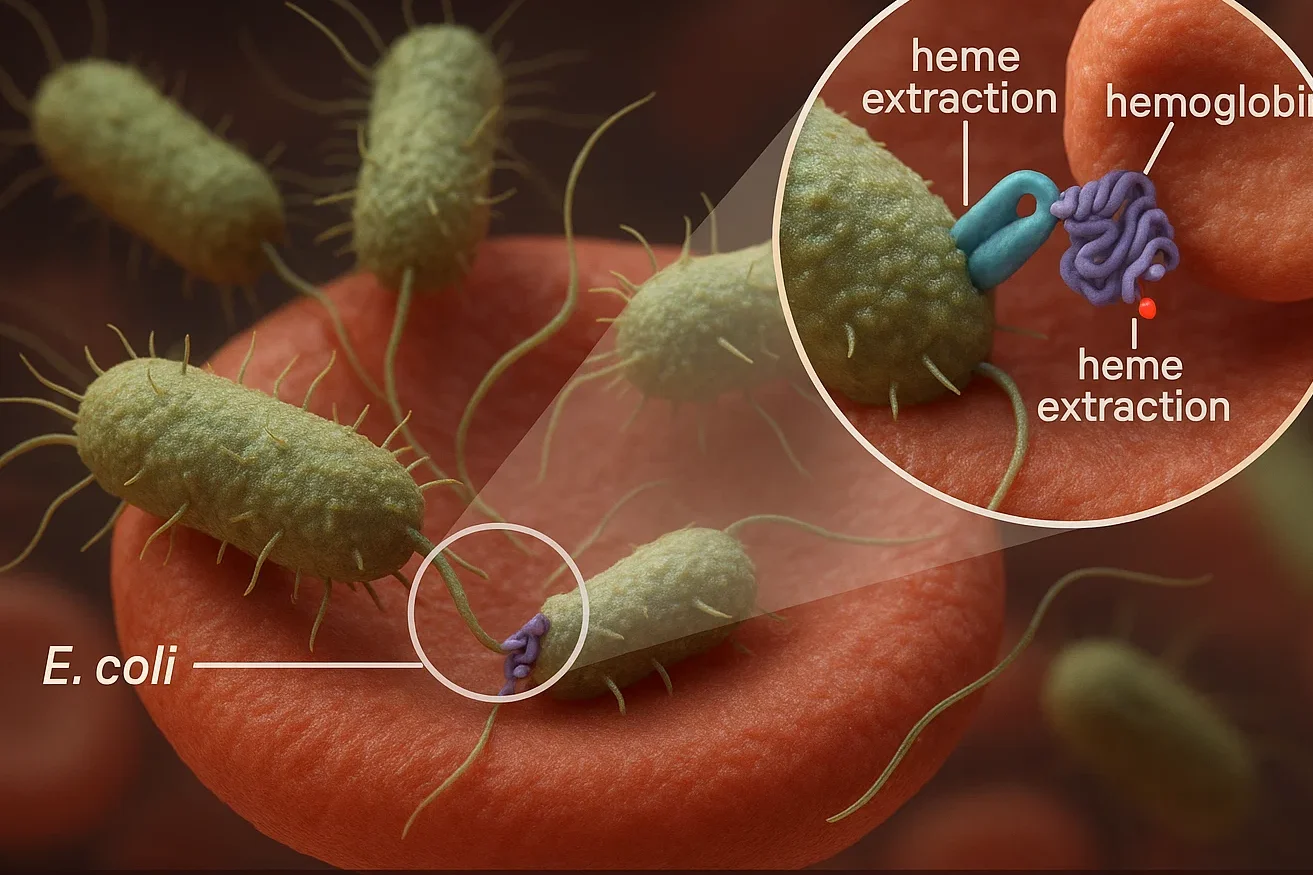
Iron is one of the vital parameters that bacteria require to survive and grow when the infection takes place, as it is the fundamental nutrient in many biological activities. The human body however keeps iron bound up and sequestered as part of its defenses, a strategy dubbed nutritional immunity that prevents the availability of free iron to be taken up by bacteria.
What Is Heme Piracy and Why Does It Matter?
Numerous disease-causing bacteria, such as E. coli and its close cousin, Shigella, have developed methods of stealing iron in the host; they do this by stealing heme, which is an iron-containing molecule in hemoglobin (the oxygen carrying protein in red blood cells). The term heme piracy is applied to this process.
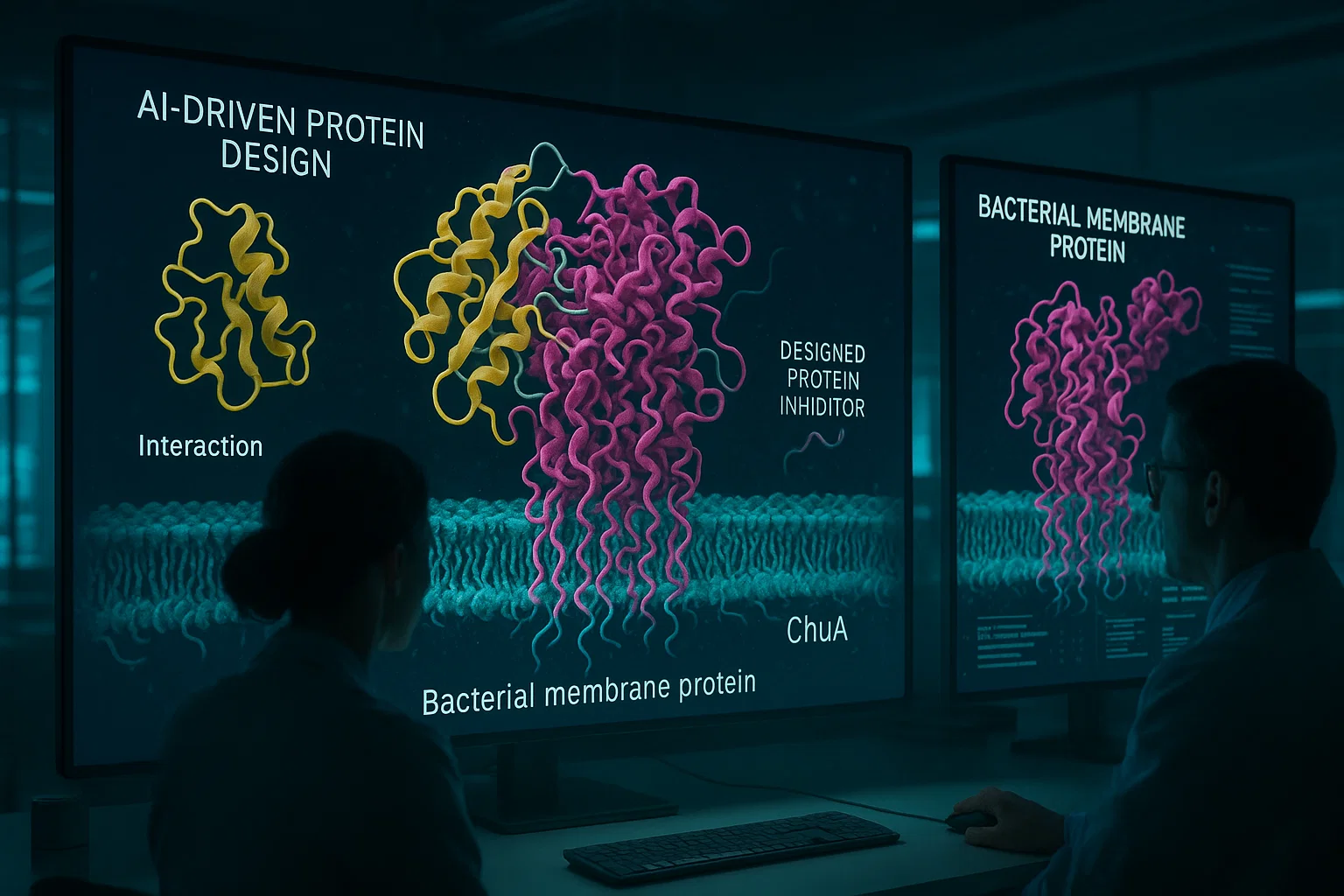
E. coli employs a dedicated outer membrane protein, named ChuA, to bind hemoglobin on the host and then very quickly depletes it of heme. This heme is imported by the bacteria to serve its iron needs that are necessary in the survival and virulent process of the bacteria.
Inhibition of this process may therefore deprive bacteria of iron to grow and act pathogenically.
The Breakthrough: Designing Proteins to Prevent Heme Uptake
A team of researchers, including Daniel R. Fox and Rhys Grinter, has managed to bring together state of the art technologies such as structural biology, artificial intelligence guided protein design, and microbiology to target the ChuA protein and starve pathogenic E. coli cells of the heme nutrient.
How They Made It
They initially determined the structure and the operating mechanism of ChuA such as the method through which hemoglobin is bound as well as the concept through which heme is released.
With the help of the powerful AI structure prediction and design programs AlphaFold2 and RFdiffusion, they created thousands of candidate structures of proteins that could act as what is known as a binder, attaching themselves to ChuA and preventing it binding hemoglobin.
The team used these proteins to synthesize in the lab and test their capacity to inhibit the growth of bacteria when the sole source of iron was in form of hemoglobin.
Key Findings: Effective Inhibition of E. coli Growth
Several de novo-designed proteins were found to strongly inhibit the growth of E. coli by preventing ChuA from extracting heme from hemoglobin, even at very low (nanomolar) concentrations.
Discovery of these structured, designed proteins, by X-ray crystallography and cryo-electron microscopy (cryo-EM), further supported that these created proteins bound ChuA with intended precision on the apparent hemoglobin binding site.
Importantly, these inhibitors did not affect the uptake of free heme, indicating a highly specific mode of action targeting the heme piracy mechanism.
Why This Is a Game-Changer in Antimicrobial Strategies
The conventional antibiotics kill bacteria or stop their development but in the long term bacteria develop resistance to conventional antibiotics. This is a new strategy to attack bacterial nutrient acquisitions which is a new therapeutic model that does not kill the bacteria but deprives them of vital nutrient uptakes.
The AI-based protein design process has great potential in the future in regards to the creation of highly specific proteins that inhibit membrane proteins such as ChuA due to the fact that:
It allows designing molecules that are capable of targeting the flexible and dynamic segments of the proteins which are not accessible to the small molecules utilized traditionally.
It enables a candidate protein to be generated and optimized very quickly.
It could be modified to be used against other bacterial pathogens that share such systems to acquire nutrients.
Possible applications and Future Advancements
The proteins that were designed within the current work give a validation proof of concept to new antimicrobial compounds, which can be exploited on a standalone basis or as a combination strategy, including known antibiotics.
The fact that ChuA is critical to bacterial virulence, but absent in human cells means the target is very specific thus reducing side effects.
Further studies can be based on the enhancement of the stability and clinical delivery of these protein inhibitors.
Another application of this working procedure is inhibitor design of other important membrane proteins in bacterial nutrient uptake or pathogenesis.
Conclusion: New Anti-Drug Weapons Against Drug-Resistant Bacteria
The original contribution of Fox, Grinter, and al. is a potential breakthrough in the war against resistance to anti-E. coli antibiotics. They have utilized the power of AI protein design to develop specific inhibitors to inhibit, in a rather efficient way, the capacity of the bacteria as able to steal iron form their host, a key step in facilitating the infection.
This deprivation of nutrition approach holds great potential to create new, specific treatment modalities against pathogenic micro-organisms that are smarter and thus better able to overcome resistant organisms and end the debate about patient outcomes in this area.
Reference:
Fox, D.R., Grinter, R., et al. (2025). Inhibiting heme piracy by pathogenic Escherichia coli using de novo-designed proteins. Nature Communications.